paired end sequencing wikipedia
The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phix-musigmaxLp0 where mu is the mean sigma. Paired-end sequencing facilitates detection of genomic.

What Is Mate Pair Sequencing For
Library preparation protocols -- In short PE protocols attach an adapter.

. Pairs come from the ends of the same DNA strand. Introduction to Mate Pair Sequencing. Paired end sequencing refers to the fact that the fragment s sequenced were sequenced from both ends and not just the one as was true for first generation sequencing.
RRNA depletion Recommended Sequencing Depth. The method uses rolling circle replication to. For those not familiar with paired-end reads check out this post.
Thirty-six or 50 bp reads are sufficient to identify most chromatin interacting pairs using Illumina paired-end sequencing. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. 10-20M paired-end reads or 20-40M reads RNA must be high quality RIN 8 Total RNA.
Shallow Whole Genome Sequencing shallow WGS also known as low pass whole genome sequencing is a new and high-throughput technology to achieve genome-wide genetic. In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a.
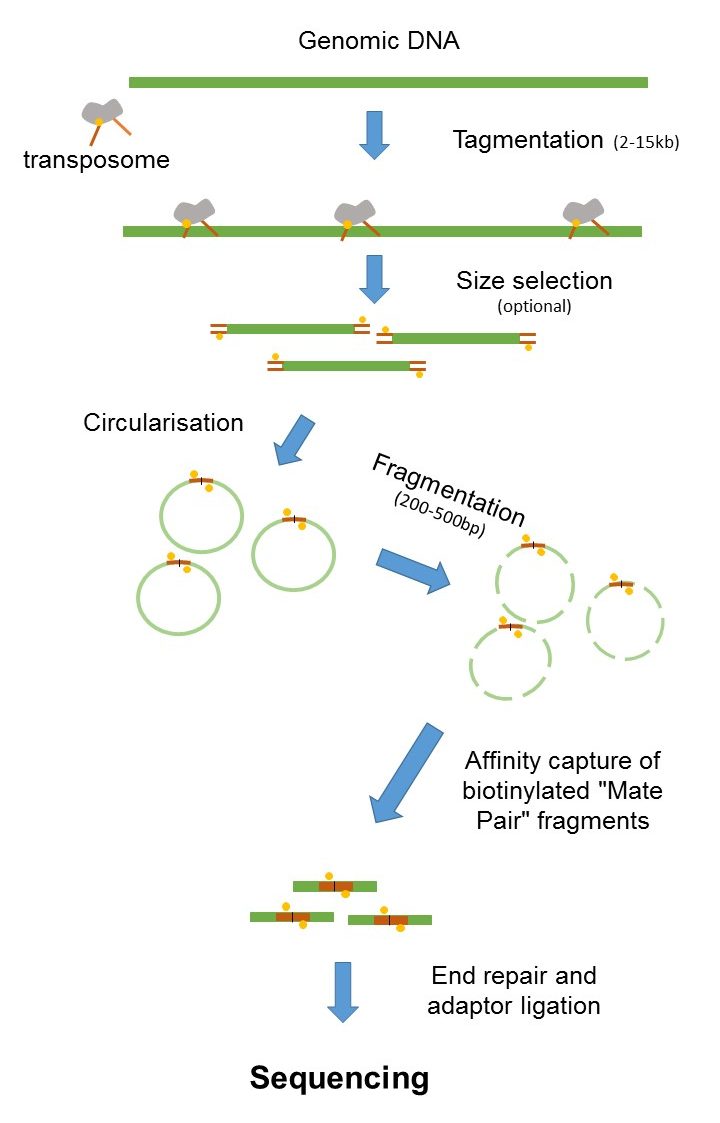
The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. The figure shows the. It has very nice and simple illustrations along with explanations on the terminology used in paired-end.
There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter. The differences between PE and MP reads include. Preparing Samples for Paired-End Sequencing Sample Preparation Kit Contents Check to ensure that you have all of the reagents identified in this section before proceeding to sample.
Read 1 often called the forward read extends from. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of.
DNA nanoball sequencing is a high throughput sequencing technology that is used to determine the entire genomic sequence of an organism. Since the average size of fragments in the library is 250 bp 50bp. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including.

Bacterial Chromosome Replication Chromosome Biology Microbiology

What Are Paired End Reads The Sequencing Center
Plos Computational Biology Informatics For Rna Sequencing A Web Resource For Analysis On The Cloud

Paired End Shotgun Sequencing Okinawa Institute Of Science And Technology Graduate University Oist
Mate Pair Sequencing France Genomique

Bacterial Chromosome Replication Chromosome Biology Microbiology
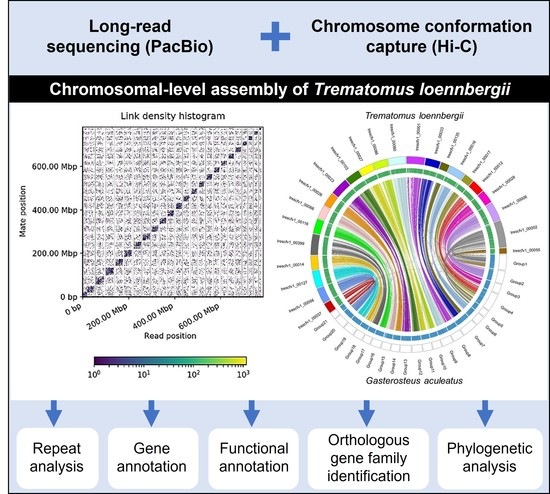
Diversity Free Full Text Chromosomal Level Assembly Of Antarctic Scaly Rockcod Trematomus Loennbergii Genome Using Long Read Sequencing And Chromosome Conformation Capture Hi C Technologies Html

What Is Mate Pair Sequencing For

Continents And Oceans Ks1 Lesson Plan Activities Teaching Resources Continents And Oceans World Map Continents Continents

Illumina Paired End Sequencing Youtube

Pin On Teaching Asian American P I History

What Is The Difference Between Single End And Paired End In Sequencing






